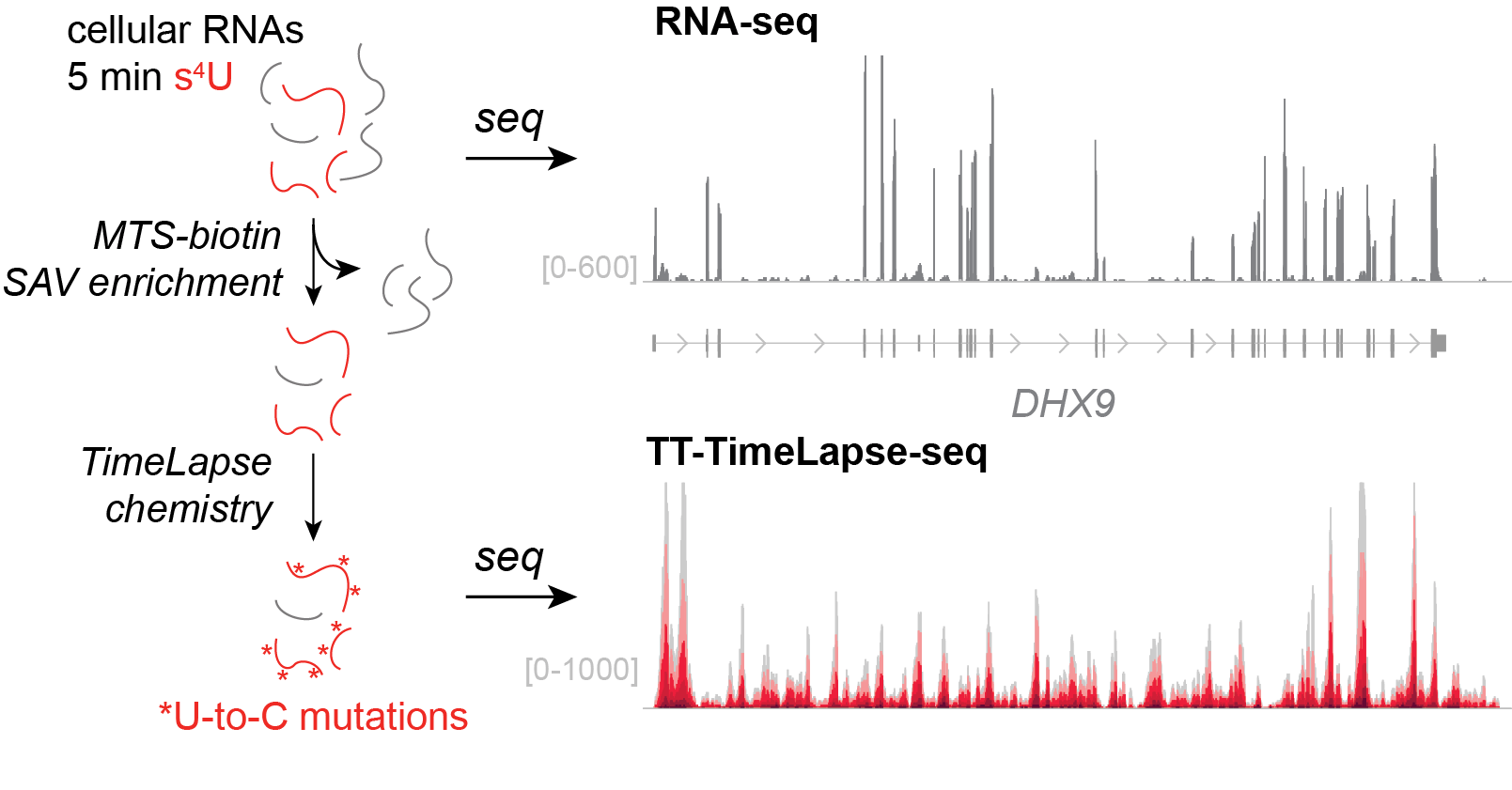
TT-TimeLapse-seq. Many RNAs are transcribed and degraded almost immediately. Because of the high rates of degradation, the steady-state concentrations of these RNAs are very low. Others have developed very short metabolic labeling protocols, including TT-seq, that allow these transient RNAs to be labeled with s4U and captured through biochemical enrichment. We have improved on these approaches through the development of TT-TimeLapse-seq. This protocol makes two improvements. First, using MTS-biotin, we can more efficiently capture s4U-labeled RNA. Second, using TimeLapse chemistry, we can identify sites where s4U was incorporated, allowing us to distinguish bona fide newly made RNAs from biochemical contaminants.