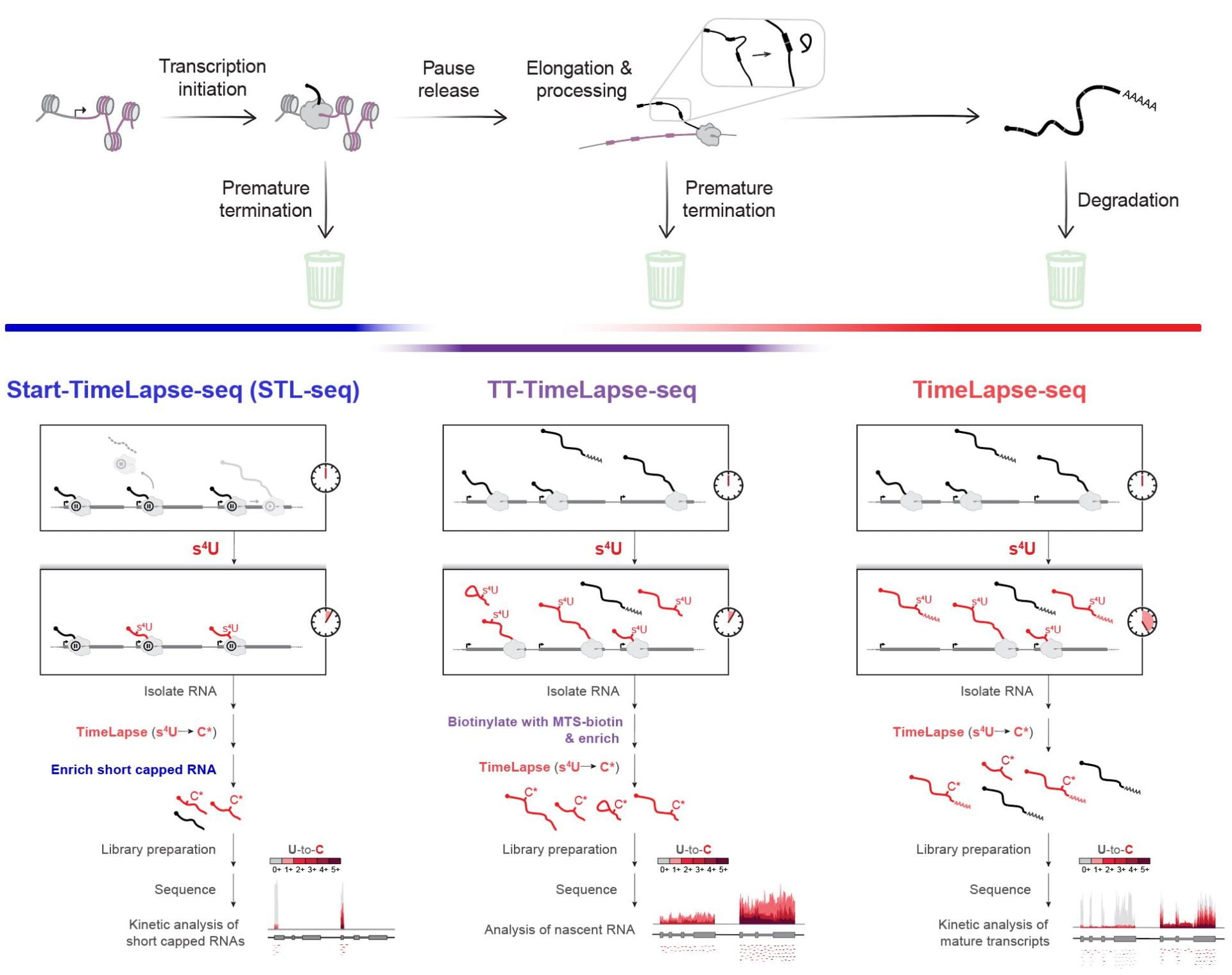
The development and application of methods to track the dynamics of cellular RNAs. Our work has contributed to our understanding of RNA population dynamics. RNAs are continuously transcribed and degraded, yet methods such as RNA-sequencing provide only a snapshot of RNA abundance. To reveal the rich regulated dynamics of the transcriptome, we have developed chemical approaches that improve on the use of 4-thiouridine metabolic labeling (Duffy et al., Mol. Cell, 2015). Through collaboration, we used this improved disulfide chemistry to study the role of m6A modification in the immune response (Li et al., Nature, 2017). We have developed a nucleoside recoding strategy to convert s4U to cytosine analogues and s6G to adenine analogues (Schofield et al., Nat. Methods, 2018; Kiefer et al., J. Am. Chem. Soc., 2018). This mutational mapping approach allows analysis of RNA population dynamics without the need for biochemical enrichment. Our work has uncovered isoform-specific differences in the turnover of a disease related transcript that encodes for the polycomb/trithorax protein, ASXL1. Most recently, we combined TimeLapse chemistry with Start-seq to probe the kinetics of transcription initiation and promoter proximal pausing in metazoa (Zimmer et al., Mol. Cell, 2021). Start-TimeLapse-seq (STL-seq) revealed the principles of transcriptional regulation at the pause site and provides the first approach to measure TSS-specific pausing kinetics in a non-perturbing manner.
Combined, our three metabolic labeling methods allow us to probe the kinetics of transcription and RNA across a transcript’s entire life cycle. From transcription initiation to degradation of the mature transcript, one or multiple of our methods can provide quantitative kinetic information about each step of the RNA synthesis pathway.
Duffy E.E., Rutenberg-Schoenberg M., Stark C.D., Kitchen R.R., Gerstein M.B., and Simon M.D. (2015) Tracking distinct RNA populations using efficient and reversible covalent chemistry. Mol Cell, 59(5), 858-66. PMID: 26340425.
Schofield J.A., Duffy E.E., Kiefer L., Sullivan, M.C. and Simon M.D. (2018) TimeLapse-seq: Chemistry to Add a Temporal Dimension to RNA-seq. Nat Methods, doi:10.1038/nmeth.4582. PMID: 29355846
Kiefer L., Schofield J.A. and Simon M.D. (2018) Expanding the Nucleoside Recoding Toolkit: Revealing RNA Population Dynamics with 6-thioguanosine. J. Am. Chem. Soc., doi: 10.1021/jacs.8b08554
Zimmer J.T., Rosa-Mercado N.A., Canzio D., Steitz J.A., and Simon M.D. (2021). STL-seq reveals pause-release and termination kinetics for promoter-proximal paused RNA polymerase II transcripts. Mol Cell. doi: 10.1016/j.molcel.2021.08.01